Filters: do only what is needed to be done!
As mentioned already several times, the main role of the database is to support the processor execution in providing the input items and in storing the output products. Not everything can be efficiently stored in a database, for example large chunk of binary data are better saved to the disc, in this case, the database will know the path where these data are stored.
One advantage of the database is that you can apply selection rules and you do not have to process the whole dataset if you do not need it. To help you in this, MAFw is offering you a ready-to-use solution, the db_filter.
This module is providing a filtering system that allows users to precisely control which data is processed by each processor. This system offers several key benefits:
Precision Control: Users can define exactly which records should be included or excluded from processing
Performance Optimization: By filtering early in the pipeline, unnecessary computations are avoided
Flexibility: Supports various filtering operations including simple conditions, logical combinations, and conditional logic
Configuration-Driven: Filters can be easily configured through the steering files without code changes
Scalability: Complex filtering scenarios can be expressed through logical expressions and nested conditions
The filtering system is built around three main components plus an extra one:
ProcessorFilter- For managing multiple model filters in a processor
ModelFilter- For defining filters at the model level
ConditionNode- For defining filters at the field level
ConditionalFilterCondition- For conditional filtering logic (this is the extra)
and they implement the following hierarchical structure:
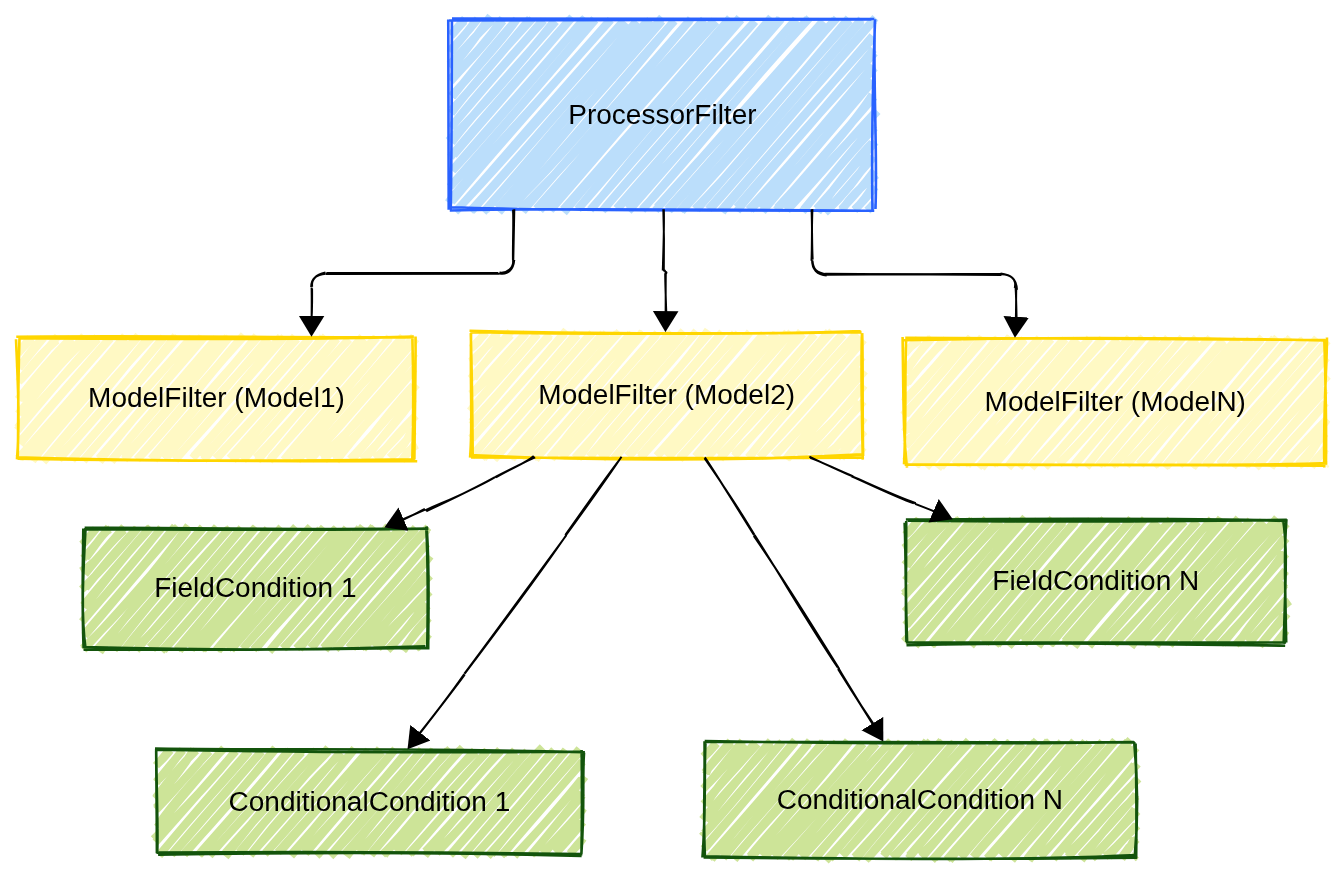
Fig. 7 The description of the filtering hierarchical structure.
How to use a filter
The filter native playground is in the implementation of the get_items() method and of course it can only work if the list of items is retrieved from the database.
Let us assume that our Processor, named AdvProcessor, is using three models to obtain the item lists. Everything is nicely described in the ERD below. The three models are interconnected via foreign key relations. There is a fourth model, that is where the output data will be saved.
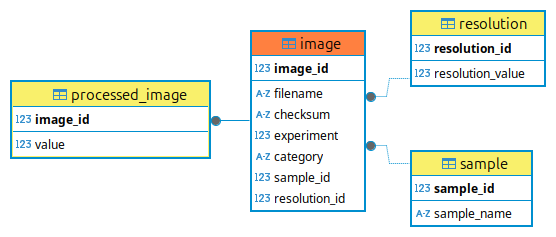
Fig. 8 The ERD of the AdvProcessor.
The real core of our database is the image table, where our data are first introduced, the other two on the right, are kind of helper tables storing references to the samples and to the resolution of our images. The processed_image table is where the output of our AdvProcessor will be stored.
To realize this database with our ORM we need to code the corresponding model classes as follows:
1class Sample(MAFwBaseModel):
2 sample_id = AutoField(primary_key=True, help_text='The sample id primary key')
3 sample_name = TextField(help_text='The sample name')
4
5class Resolution(MAFwBaseModel):
6 resolution_id = AutoField(primary_key=True, help_text='The resolution id primary key')
7 resolution_value = FloatField(help_text='The resolution in µm')
8
9class Image(MAFwBaseModel):
10 image_id = AutoField(primary_key=True, help_text='The image id primary key')
11 filename = FileNameField(help_text='The filename of the image', checksum_field='checksum')
12 checksum = FileChecksumField(help_text='The checksum of the input file')
13 experiment = IntegerField(default=1,
14 help_text='A flag for selection of the experiment. Flags are bitwise combinable')
15 category = TextField(
16 help_text='A text string to describe the image category. Accepted values are: "STANDARD", "SUT", "REFERENCE"')
17 sample = ForeignKeyField(
18 Sample, Sample.sample_id, on_delete='CASCADE', backref='sample', column_name='sample_id'
19 )
20 resolution = ForeignKeyField(
21 Resolution, Resolution.resolution_id, on_delete='CASCADE', backref='resolution', column_name='resolution_id'
22 )
23
24class ProcessedImage(MAFwBaseModel):
25 image = ForeignKeyField(
26 Image,
27 Image.image_id,
28 primary_key=True,
29 column_name='image_id',
30 backref='raw',
31 help_text='The image id, foreign key and primary',
32 on_delete='CASCADE',
33 )
34 value = FloatField(default=0)
By now, you should be an expert in ORM and everything there should be absolutely clear, otherwise, take your chance now to go back to the previous sections or to the peewee documentation to find an explanation. Note how the Image class is making use of our FileNameField and FileChecksumField. We added also a bit of help text to each field, in order to make even more evident what they are.
Particularly interesting is the experiment field in the Image model. This is a binary flag and can be very useful to assign one file (an image in this case) to one or more experiments. For example, imagine you have three different experiments in your data analysis; you assign to the first experiment the label 1 (binary: 0b1), to the second the label 2 (0b10) and to the third the label 4 (0b100). Now, if you want an image to be used only for experiment 1, you set the experiment column to 1; similarly if you want an image to be part of experiment 1 and 3, then you set its experiment column to 1 + 4 = 5 (b101). In fact, if you bit-wise AND, this image with the label of the experiments (5 BIT_AND 1 = 5 BIT_AND 4 = True) you will get a True value.
For each foreign key field, we have specified a backref field, so that you could get access to the related models. Pay attention also at the highlighted lines, where we define foreign key fields to other tables. Peewee follows Django style references, so actually the field object is named with the noun of the object you are referring to. This will allow the following:
image.sample # Resolve the related object returning a Sample instance, it costs an additional query
image.sample_id # Return the corresponding Sample's id number
The primary source of input is the Image; however, you may wish to process only images that meet specific criteria, such as belonging to a particular sample or being captured at a certain resolution. Unfortunately, this information is not explicitly included in the Image model. Only the resolution_id and the sample_id are included in the image table: those primary keys are excellent for a computer, but for a human being it is better to use sample names and resolution values. The solution is to use a join query in order to have all fields available and then we will be able to apply the configurable filters from the TOML steering file to limit the selection to what we want.
Since we have three input models for our processor we can have up to three different Model level filters defined in the steering files. The processor is storing all the filters configured in the steering files in a container class filter register that will be the main actor of our get_items() implementation that follows:
1def get_items(self):
2
3 # first of all, let us be sure that the tables exist
4 # the order is irrelevant, the ORM will find the best creation strategy.
5 # if the table already exists, nothing will happen.
6 self.database.create_tables([Sample, Resolution, Image, ProcessedImage])
7
8 # if you want to remove widow rows from the output table or verify the checksum do it now!
9 remove_widow_rows([Image, ProcessedImage])
10
11 # did we select new_only in the global filter?
12 # if yes, prepare an additional condition in which we specify that the
13 # Image.image_id should not be among the image_id of the ProcessedImage.
14 # if no, then just accept everything.
15 if self.filter_register.new_only:
16 # let us get a list of all already processed items.
17 # since the item in the output table are stored using the same primary key,
18 # this will simplify the discrimination between new and already processed items.
19 existing_entries = ProcessedImage.select(ProcessedImage.image_id).execute()
20
21 # build the condition
22 existing = ~Image.image_id.in_([i.image_id for i in existing_entries])
23 else:
24 existing = True
25
26 # finally let us make the query.
27 query = (Image.select(Image, Sample, Resolution)
28 .join(Sample, on=(Image.sample_id == Sample.sample_id), attr='s')
29 .switch(Image)
30 .join(Resolution, on=(Image.resolution_id == Resolution.resolution_id), attr='r')
31 .where(self.filter_register.filter_all())
32 .where(existing)
33 )
34 return query
The processor filter register comes up in two points (highlighted in the code), when can retrieve a boolean flag representing whether we want to process input data for which an output record already exists or not.
The second usage is in the query, where we generate a peewee combined expression representing the whole combination of all user defined (and configured) filters. Let’s move on and see how we configure a filter!
How to configure a filter
In a steering file, there is a table for each processor where you can set the configurable active parameters. To this table, you can add a sub-table named __filter__, containing other tables, one for each input Model. The reason for the underscores is to avoid to pollute the processor parameter namespace. This is how it will look like:
[AdvProcessor]
param1 = 15
[AdvProcessor.__filter__.Resolution]
resolution_value = 25
[AdvProcessor.__filter__.Sample]
sample_name = 'sample_00*'
In the example above, AdvProcessor has two *.__filter__.* tables, one for Resolution and one for Sample. When the steering file will be parsed, the processor constructor will automatically generate two filters: for Resolution it will put a condition that the resolution field must be 25 and for Sample, the sample_name column should be ‘sample_1’. You could add a third table for the Image model, if you wish.
In this way, the processor initialization machinery will create the necessary filters and add them to the processor filter register from where we can retrieve it.
The new_only parameter of the register can also be configured in the steering file. By default, you can find in the steering file in the general section a parameter new_only = true. This flag will be applied to all processors in the pipeline, but you can change it for each processor in this way:
[AdvProcessor]
__new_only__ = false
param1 = 15
# The rest as before...
In this way, for the AdvProcessor the new only flag will be False [1], while the rest is left to the default value.
If we use this configuration the AdvProcessor will obtain an input item list with only the input images having a resolution_value of 25 AND a sample_name matching the glob style string sample_00*.
It is important to underline that the filters on the models are joined (by default) using an AND. In this simple example we had one field for each model being the subject of the filter, but you could have as many as you like. Inside a model, the default joining logic is again AND.
The configuration of these filters is done using the so called single value approach, even if for the sample_name we specified an * in the name. The reason is that we have assigned with the = symbol one single value to the model field.
The filter will interpret the meaning of = differently depending on the type of the right hand side value, following this table:
Filter field type |
Logical operation |
Example |
|---|---|---|
Numeric, boolean |
== |
Field == 3.14 |
String |
GLOB |
Field GLOB ‘*ree’ |
List |
IN |
Field IN [1, 2, 3] |
Explicit operation configuration
In the previous example we have seen how to select one specific field to be exactly equal to a given value, but maybe our goal is to select an interval, or performing a bitwise logical operation. The filter system also supports explicit operation specification, allowing you to define the exact logical operation to be applied. Here is an example:
[AdvProcessor]
param1 = 15
[AdvProcessor.__filter__.Resolution]
resolution_value = {op = ">=", value = "25"}
[AdvProcessor.__filter__.Image]
experiment = { op = "BIT_AND", value = 5 }
In this configuration example, you can see to practical usage of the so-called explicit operation configuration. It is called explicit, because this time you clear specify which operation you want to apply. So, in this case we will be getting all images with a resolution value greater or equal to 25 and an experimental flag returning true when bitwise AND’ed to 5. If you remember, in our introduction, this is equivalent to select experiment labelled with 1 and with 4.
The supported operations include [2]:
Operation |
Description |
Example |
|---|---|---|
== |
Equal |
Field == 42 |
!= |
Not equal |
Field != 42 |
< |
Less than |
Field < 100 |
<= |
Less than/equal |
Field <= 100 |
> |
Greater than |
Field > 0 |
> |
Greater than/equal |
Field >= 10 |
GLOB |
Pattern matching |
Field GLOB ‘test*’ |
LIKE |
SQL LIKE |
Field LIKE ‘test%’ |
REGEXP |
Regular expression |
Field REGEXP ‘^[A-Z]’ |
IN |
Value in list |
Field IN [1, 2, 3] |
NOT_IN |
Value not in list |
Field NOT_IN [1, 2] |
BETWEEN |
Value between |
Field BETWEEN [1, 10] |
BIT_AND |
Bitwise AND |
Field & 5 != 0 |
BIT_OR |
Bitwise OR |
Field | 7 != 0 |
IS_NULL |
Field is NULL |
Field IS NULL |
IS_NOT_NULL |
Field is not NULL |
Field IS NOT NULL |
Note
The default sqlite library provides only an abstract definition of the regular expression matching. In simple words, it means that the user needs to implement the user function regexp(), or using any sqlite extensions that implements it.
In summary, if you are using the vanilla sqlite, you cannot use the REGEXP operator in your filter and you need to reformulate your filtering condition using a combination of other string matching tools.
Logical Composition
In the examples seen so far, the field conditions inside of a ModelFilter were logically AND’ed to generate the condition for that specific model. If more model filters are present, then the overall processor filter was generated logically AND’ing all model filters. Always keep in mind this hierarchical structure: Field Condition < Model Condition < Processor Condition described in Fig. 7.
You may want to have a different logical composition, defining how field conditions inside a model or model filters inside a processor are combined. To do so use the __logic__ keyword to define the logical composition. See the example below.
[AdvProcessor]
param1 = 15
__logic__ = "(Resolution OR Sample) AND Image"
[AdvProcessor.__filter__.Resolution]
resolution_value = {op = ">=", value = "25"}
[AdvProcessor.__filter__.Image]
__logic__ = "category OR image_id"
experiment = { op = "BIT_AND", value = 5 }
category = "beautiful_images"
[AdvProcessor.__filter__.Image.image_id]
__logic__ = "good_images AND NOT bad_images"
good_images = {op = "BETWEEN", value = [100, 145]}
bad_images = {op = "IN", value=[105, 109]}
[AdvProcessor.__filter__.Sample]
sample_name = "sample_00*"
sample_id = [1,2]
This is a very advanced configuration and I hope you will appreciate how easy is to obtain.
First of all there is a logic sentence in the AdvProcess table. It says that we want to combine the expressions for the Resolution and the Sample models using OR. The result of this should be combined with AND with the expression for the Image model.
The resolution filter is an explicit condition as seen in the previous paragraph. There is only one condition for this model, so nothing to combine. For the Sample model, we have two field conditions and no __logic__ keyword, so the two conditions will be combined with the default AND.
The filter on Image is a bit more complicated. First of all, it has a logic statement that will be used to combine the field condition. The statement is mentioning category and image_id, but nothing about experiment, as a consequence the condition on the experiment will be simply ignored. The category condition is a simple value one while the one over image_id is a nested. In fact for image_id, we have an additional table describing what we want. This is a combination governed by a logic statement where we have defined two sub-conditions, a range of good images to be combined with some bad images.
This whole configuration is equivalent to this SQL statement:
SELECT
*
FROM
image
join resolution using(resolution_id)
join sample using(sample_id)
WHERE
( resolution.resolution_value >= 25 OR
(sample.sample_name GLOB "sample_00*"
AND sample.sample_id IN (1, 1) ) AND
( image.image_id BETWEEN 100 and 145 AND NOT image.image_id IN (105, 109))
Summary
There are three different filtering levels: Field, Model, Processor.
For each level, you can define as many conditions as you wish. Those conditions are by default combined using AND, but more advanced logical composition can be achieved using the __logic__ keyword via a simplify grammar.
The logical grammar uses only three logical operators: AND, OR and NOT. The variable names are the model names at the processor level, the field name at the model level and the condition name at the field level.
Be aware that the grammar is case sensitive: the operators must always be written in capitol letters, while the variable names should respect the original case. Use parenthesis to specify the order. When a logic statement is provided, variables that are not mentioned in the statement will not be included in the filter generation.
Each condition lives in its own scope and cannot be directly linked to conditions in other scope. So for example, field conditions of defined in Resolution cannot be included in the logic statement of the Image model.
Conditional filters
In the introduction, we mention the three pillars on which the filtering system is based plus one extra. This extra one is the conditional filter! They allow expressing logic where one field’s criteria depend on another field’s value.
They live inside the model scope since they relate different fields of the same table. For each model, you can have as many as you like and you can also include them in the model level logic statement.
[AdvProcessor]
param1 = 15
[AdvProcessor.__filter__.Image]
__logic__ = "resolution_id AND conditional_1"
resolution_id = 1 # this is the foreign key value
[[AdvProcessor.__filter__.Image.__conditional__]] # this is a list, note the double []
name = 'conditional_1'
condition_field = 'category'
condition_op = 'GLOB'
condition_value = 'beautiful'
then_field = 'raw_image_id'
then_op = 'BETWEEN'
then_value = [100, 140]
else_field = 'raw_image_id' # OPTIONAL
else_op = '>' # OPTIONAL
else_value = 200 # OPTIONAL
The conditional filter translates as follow: if the category is matching ‘beautiful’, then select raw_image_id between 100 and 140, otherwise (if not matching ‘beautiful’) select raw_image_id greater than 200.
The SQL equivalent is:
WHERE resolution_id = 1 AND
(( category GLOB "beautiful" AND raw_image_id BETWEEN 100 AND 140 )
OR ( category NOT GLOB "beautiful" AND raw_image_id > 200 ))
Summary
Conditional filters allows to select one field based on the value of another fields.
You can add as many as you like, just repeat the same table header (with the double set of []).
How to use the new_only filter
Basic usage example
The basic usage of the new only filter is based on the relation between input and output table. Going back to our previous example, we have an Image model containing our input data and a ProcessedImage model containing the output of the processor. The two models are in a 1-to-1 relation via a foreign key constrain linking the primary keys of both models. In this case the selection of the so-called new_only elements, it is to say the items in the input list for which an output is not yet existing is very easy to obtain. You get the whole list of primary keys in the output table (this is the existing list) and you compare with the list of primary keys in the input table. If the two lists are different, then run the processor only on the difference!
Note
The output primary key list is at most equal to the input primary key. It cannot contain elements that are not in the input table by definition of 1-to-1 relation.
Programmatically you can obtain this in the implementation of the Processor.get_items() method as shown in this code snippet at lines 19 and 22.
New only flag and processor parameters
Let’s now consider another common scenario where the 1-to-1 relation between two tables are not enough. You have implemented a Processor that is responsible to perform a gaussian filtering of your data and for this you have defined gauss_sigma as an ActiveParameter. You run the pipeline for the first time and your GaussFilter is doing its job using gauss_sigma = 3, but then you realize that 3 is probably too much and you want to lower it down to say 2. You change the steering file and you re-run it and very likely nothing will happen. The reason is that if you have implemented the new_only filter in the get_items() as shown before, the output table is already containing all the filtered items from the input table.
A trick to force the re-processing is to delete the items manually from the database or one of its output files (if any and if you have included the remove_widow_db_rows() or verify_checksum()) but this is not really comfortable. The most elegant solution is to include a column in the output table to store the value of gauss_sigma and then adding a where condition in the query looking for the existing items.
Look at this query snippet example:
def get_items(self):
# add here all your checks, table creations, filter bindings
if self.filter_register.new_only:
query = (
Image.select(OutputTable.input_id)
.where(OutputTable.gauss_sigma == self.gauss_sigma)
)
existing = ~InputTable.input_id.in_([row.input_id for row in query])
else:
existing = True
items = (InputTable.select(InputTable)
.where(self.filter_register.filter_all())
.where(existing)
)
return items
This approach is very handy, because it allows to link the entries in the database with the parameters set in the steering file, but it must be used with care, because changing a parameter will trigger the reprocessing of all entries while you might be thinking that this will only apply to the added items only.
Advanced new_only filtering with multiple column primary keys
Before moving on to the next section, we would like to show you another implementation tip where multiple column primary keys and new_only filtering work nice together. In the basic usage the new only flag the input and output tables were linked by a foreign key based on a single column primary key. Now let’s see how we can handle the situation where the input table is the cross join of two tables with different primary keys (image_id from ProcessedImage and method_id from CalibrationMethod) and the output table CalibratedImage has the combined primary key (image_id, method_id). The models described here refers to the ones described here.
The idea is to use field combinations and for this you can rely on some helper functions defined in the db_tools module. Let’s have a look at a possible implementation.
1def get_items(self):
2
3 # check if we want to process only new items
4 if self.filter_register.new_only:
5 # get a combined primary key because the output table (CalibratedImage) has
6 # a multicolumn primary key
7 existing_combo = combine_pk(CalibratedImage,
8 alias_name = 'combo',
9 join_str = ' x ')
10
11 # execute a select on the output table for the combined field
12 existing_entries = CalibratedImage.select(existing_combo).execute()
13
14 # to find the existing entries in the input model, we need to combine the fields
15 # and compare them to the list of combined pks from the target model.
16 # Pay attention to two things:
17 # 1. In the where conditions you cannot have an alias, so the
18 # combine_fields is not aliased.
19 # 2. The argument of the in_ must be a python list and not the pure select.
20 existing = ~combine_fields(
21 [ProcessedImage.image_id,
22 CalibrationMethod.method_id],
23 join_str = ' x '
24 ).in_([entry.combo for entry in existing_entries])
25 else:
26 existing = True
27
28 query = (
29 ProcessedImage.select(ProcessedImage, Image, Sample,
30 Resolution, CalibrationMethod)
31 .join_from(ProcessedImage, Image, attr='_image')
32 .join_from(Image, Sample, on=(Image.sample_id == Sample.sample_id), attr='_sample')
33 .join_from(Image, Resolution, on=(Image.resolution_id == Resolution.resolution_id), attr='_resolution')
34 .join_from(ProcessedImage, CalibrationMethod, on=True, attr='_calibration_method')
35 .where(existing)
36 .where(self.filter_register.filter_all())
37 )
38 return query
The code snippet is rather self-explanatory and well commented. The most important parts are the lines 7-9: they are the SQL equivalent of:
SELECT image_id | ' x ' | method_id FROM calibrated_image
so it will return one single column of text where each row is something like 1 x 1, 2 x 1….. You can change the joining string to whatever you like most, the only requirement is that it must be the same as the one used at line 23.
The next step is to build a query of all possible combinations of the two fields ProcessedImage.image_id and CalibrationMethod.method_id. This is obtained using the combine_fields() function. Pay attention to two small details:
The output of
combine_fields()is meant to be used in the where condition and peewee does not supported aliased expression in where conditions.The in_ operator is expecting a python list as argument, so you need to transform the existing_entries query is a list with the combinations.
Why do I get an error about model binging?
It is possible that when using filters in your project, you will get an error at runtime mentioning that it was not possible to perform autobinding of the model or that the ModelFilter was no bound before it is use.
The reason for this error is very simple and the solution also straightforward. But let me fist explain what model binding means and how it is done. When you write your steering file and configure the filters, you identify the model class with a string in the TOML dictionary. The model is a class and the interpreter needs to connect (bind) the name you provide with the steering file with a Model class in order to perform the actual filters generation.
The current implementation of peewee does not provide a list or a dictionary of all the existing models in a database, but MAFw has a solution for this, the ModelRegister already introduced in the previous section Automatic model registration. If your Model inherits from MAFwBaseModel, then you are safe: the processor filter will ask the model register to provide the link between the model name and the actual class and your are good to go.
If your Models are not inheriting from MAFwBaseModel then you have to do the binding manually. It is not difficult, it is just a line of code to be added to the implementation of the get_items method just before preparing the query, Here is an example:
def get_items(self):
# create your tables, if they still do not exist
self.database.create_tables([Model1, Model2])
# now bind the filters with the models
self.filter_register.bind([Model1, Model2])
# now continue the code as in the previous examples.
Filter migration from MAFw v1.4.0
If you were using MAFw in its early version v1.4, you might be find interesting reading this paragraph. Otherwise, skip it and go to the next chapter.
There were a lot of changes to the filter module going from v1.4 to v2.0, mainly in the implementation side but some also in the interface. So if you had steering files generated with version v1.4, they probably won’t work directly with the newest version.
Here is a list of the important changes that you need to take care of:
The GlobalFilter is removed. In the old version, this was used to store the global new_only flag and, if implemented, some default filters to be applied to all processors in the pipeline. This functionality has become incompatible with the new system and has been removed. If you want to set the value of the new_only flag at the pipeline level, just use the new_only variable at the top level of the steering file and you can change it at Processor level using the __new_only__ keyword.
Filter replaced by __filter__. In the old steering files, the TOML table containing the definition of the filters for each model was identified by the word Filter (with capitol F). This has now been replaced by the keyword __filter__, with double underscores at the beginning and at the end. This is to allow you to have a processor parameter named Filter and also it is more in line with the keyword style used for the logic statement, the new only tag and the conditional filters.
Automatic binding. In version v1.4, the filter binding with the model was under the responsibility of the user, meaning that it was your task to do the binding of each filter with its model before being able to generate the filtering expression. In version v2.0, thanks to the
ModelRegisterthe binding process is happening automatically if your models inherit fromMAFwBaseModel.Missing fields in a ModelFilter. In version v1.4, if you had a field condition in a ModelFilter that was not matching any field in the bound model, the condition was silently ignored. If for example, you wanted to create a condition on a field named my_image_id and you wrote a condition line image_id = 5, the filter was not able to find the image_id field in the model and it was ignoring this statement. This behavior is incompatible with the new implementation. Now all conditions in a ModelFilter should match a field in the corresponding model. If the user provided a logic statement, not mentioning one or more conditions, they will silently ignored, otherwise the default behavior of AND’ing all of them will be applied.
What’s next
The power of filters is incredible and you can control them via the steering file without having to change a single line in your code!
The next chapter is about the library of advanced processors that MAFw is sharing to simplify your job even further. In particular, we are sure you will like a lot our plotting processors!
Footnotes